
Detection of protein targets with the Wes system is conceptually the same as conventional Western analysis. Sample proteins are fractionated either by size or charge, immobilized to a solid surface and detected using target specific antibodies.
However, the Wes system uses processes of protein fractionation, immobilization and immunodetection that are completely automated. Importantly, the technologies developed for automating these processes also make the Wes system more efficient and reproducible.
Samples containing protein targets are prepared using normal procedures with commonly used lysis buffers and reagents. Many sample types can be analyzed including lysates prepared from tissue, cultured cells or processed samples such as immunoprecipitations, GST-pulldowns or protein purification processes.
Sample preparation for the Wes run is essentially the same as for conventional westerns the samples are mixed with Wes sample master mix containing dithiothreitol, and a mixture of three fluorescent standard dyes and heat denatured. The samples are then loaded on a Wes assay plate with the other system components which include wash buffers, chemiluminescence substrate, primary and secondary antibodies. Extra wells are included in the assay plate for extra reagents, such as extra wash buffers, as needed. Once the assay plate is prepared it is placed into the Wes system with the appropriate capillary set (i.e. 13- or 25-capillary) and the run is started. The remainder of the run is completely automated including quantification.
Run data can be analyzed in two formats, as an electropherogram or as gel image (Fig. 1). The final run data can be analyzed using the Wes analysis software and which can be exported for use with other applications.
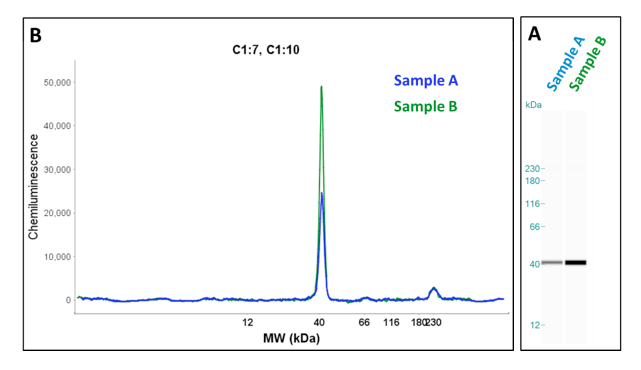
A key aspect of the Wes system is the use of capillary electrophoresis for protein separation. All the processes of protein separation, immobilization and immunodetection are accomplished in the capillary. Therefore each capillary is equivalent to an independent experiment making it possible to perform up to 24 independent experiments in a single run. This means that 24 different protein targets can be detected in one protein sample in a single run or multiple control experiments can be run simultaneously (i.e. secondary antibody alone, antigen blocking). Although this is a great advantage over conventional western blotting there is also the added power that these analyses are quantifiable and can be directly compared to each other.
The use of capillary electrophoresis also greatly reduces the sample volumes required and results in very consistent separations without effects from neighboring samples, edge effects or smiling. The capillary volume is 400nL and only requires 40nL of protein sample for each analysis. Typically, only 1 ng (0.2mg/mL) of protein is required for each capillary, however, for relatively low abundance protein targets up to 20ng (2mg/mL) of protein can be used. This is in contrast to conventional western blotting that typically requires samples containing 10ug to 200ug of protein.
The capability of using these low amounts of protein is a considerable advantage over conventional western blotting making the Wes system the better alternative when analyzing valuable samples that have limited availability.
| Lysis buffer | Vendor |
|---|---|
| RIPA | Cell Signaling, PN 9806S |
| Cell Lysis Buffer | Cell Signaling, PN 9803S |
| SDS Lysis Buffer | Millipore, PN 20-163 |
| IP Lysis buffer | Pierce, PN 87787 |
| T-Per | Pierce, PN 78510 |
| Cellytic MT | Sigma, PN C3228 |
| *Adapted from ProteinSimple publication (112611_Buffer compatibility tables R1.pdf) | |
Lysis buffer composition and protein concentration are the most important considerations when preparing samples for analysis on the Wes. The type of lysis buffer will also affect the measurement of sample protein concentration.
Molecular weight estimates are calculated using a standard curve generated for each run. Six biotinylated proteins ranging from 12 kDa to 230 kDa are used to generate the standard curve. They are prepared using the same procedure as the protein samples and their positions in the capillary are detected using streptavidin conjugated-HRP. Each protein sample in a run is then directly compared to the standard curve. This is accomplished by the inclusion of fluorescent standards in the sample loading buffer giving each sample, including the molecular weight ladder, an internal standard that is used to align the sample with molecular weight standard.
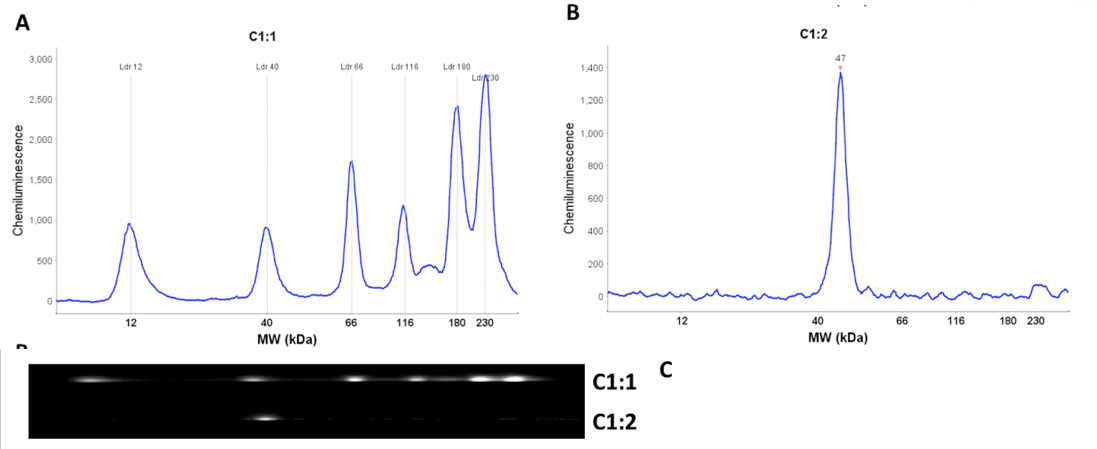
Another key technological innovation of the Wes system is the immobilization of separated proteins directly to the capillary prior to immunodetection. In conventional western blotting the sample proteins are electrophoretically transferred from the gel and immobilized onto a nitrocellulose or PVDF membrane by non-covalent interactions (i.e hydrophobic and electrostatic). The Wes system utilizes UV light catalyzed covalent crosslinking of the proteins to the capillary itself. Therefore the proteins are directly immobilized in place at the point of their migration. This completely eliminates the inefficiency associated with the process of electrophoretic transfer and non-covalent immobilization.
Immunodetection is achieved by sequential application of the primary antibodies, secondary antibodies and wash buffers to the protein conjugated capillaries. Target protein detection is accomplished by applicaton of chemiluminescence substrate to the capillary and direct capture of the light emitted during the enzyme catalyzed reaction (Fig. 2). Data is captured for predetermined time periods as defined by the user (up to seven exposure times can be defined). Having several exposure times defined gives consistent data from run to run and ensures data will be captured during the linear portion of the reaction when substrate is in excess and the signal is directly proportional to enzyme concentration. Under these conditions the signal strength is directly proportional to target protein abundance.
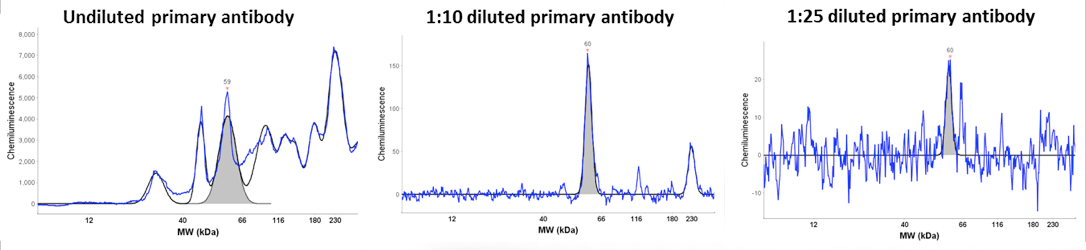
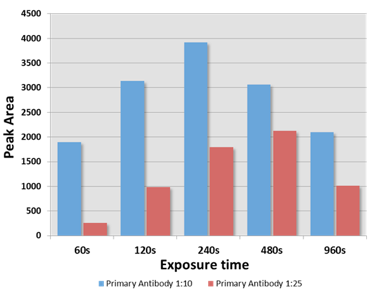
An example is given that highlights several of the advantages of the Wes system. The data is from an antibody titration experiment using an antibody that recognizes a low abundance 60 kDa protein. The Wes run was setup with equal amounts of protein lysate and detection with three dilutions of the primary antibody (undiluted, 1:10 diluted and 1:25 diluted, Figure). With increasing dilution the background signal was reduced and the specificity of the 60 kDa signal was increased with the 1:10 dilution being optimal. Analysis of the 60 kDa signal at different exposure times showed that the max signal obtained with the 120 second exposure. More importantly, this analysis demonstrated a linear increase of the signal at the 60, 120 and 240 second exposures. Moreover, the signal strength directly correlated with the antibody dilution as expected. Signal reduction with higher exposures indicates substrate depeletion.
| Cap | Peak | Name | Position | MW(kDa) | Height | Area | Width | S/N | Baseline |
|---|---|---|---|---|---|---|---|---|---|
| C1:10 | 3 | 494 | 59 | 4140.1 | 126239 | 28.6 | 190.1 | 789.1 | |
| C1:11 | 1 | 497 | 60 | 151.3 | 1899 | 11.8 | 56.3 | 171.9 | |
| C1:12 | 1 | 496 | 60 | 23.7 | 256 | 10.1 | 11.1 | 170.7 |
A subset of the sample data obtained by the Wes software (Compass) for the run is shown above. The data shown is for one exposure time and the same data for each exposure time is stored for each run performed. All of the data stored can be exported as text files or to spreadsheet applications.
Time savings is another key advantage of the Wes system over conventional immunodetection. The Wes system only requires 2.5 hours per run and since the run is completely automated the run time is exact and this time includes capturing and quantifying the data. This contrasts with a conventional western that takes at least 6-8 hours starting (typically a two day procedure) from loading sample to scanning the developed film. This offers the benefit of performing multiple runs each day providing the capability of performing 3-4 runs a day. This allows the analysis of 72 – 96 samples in a single day.
ProteinSimple antibody database. A database of qualified antibodies is maintained by ProteinSimple. Currently there are 2311 entries in the database and it is continually being updated.
The database contains the source of the antibody and the conditions that were successfully used. This includes the type of sample (i.e. cell lines, tissue), method of protein fractionation (by size or charge) and antibody dilutions. This is a great resource that can significantly decrease the amount of time that would normally be spent on developing experimental conditions.
